The speed of life
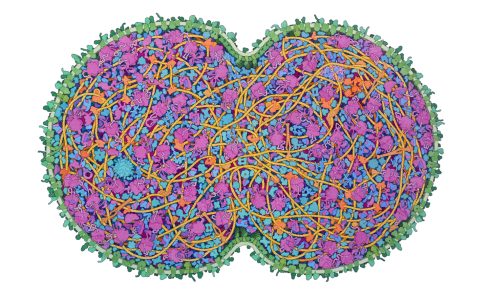
Above: A depiction of the JCVI-syn3A minimal cell, by David S Goodsell.
21 February 2025
Niko McCarthy argues that understanding the speed of cellular processes can make us better biologists
This article originally appeared in Asimov Press
There is an inverse, linear relationship between the size of a thing and the speed at which it changes. Tiny things are easier to move. Cameras may allow us to ‘slow down’ time, but they are limited by their resolution and are best suited, often, to high speeds that occur at the macroscopic scale. At the nanoscale, one must contend with both small sizes and rapid movements, pushing the boundaries of both pixels and shutter speeds. Do we really understand things that are extremely fast? I don’t think so.
Within cells, there are enzymes that perform millions of chemical reactions each second. Protein ‘motors’ make energy-storing molecules by spinning around thousands of times a minute. Sugar molecules fly by at 250 miles per hour. Every protein in the cell is hit by 1,013 water molecules every second and a protein will typically move through a cell at about 10 metres per second. (But cells are so densely crowded, and collisions so common, that this is not always their speed, merely an average.)
When I first heard these numbers, I thought they were made up. After all, how is it even possible to measure such speeds? The world’s most powerful microscope cannot ‘see’ a protein motor spinning or watch a sugar molecule move through a cell. But as a PhD student, I jumped head first into the world of biological speed. My goal was to collect some ‘fast’ numbers in biology and understand the experiments that brought them to light. My search made me appreciate how remarkable it is that life functions at all considering the chaotic conditions in which cells exist. It also gave me a new appreciation for biology and the incredible exactitude that one must have to engineer it – let alone engineer it successfully.
 Within cells, enzymes catalyse millions of chemical reactions each second. But which processes limit the speed at which cells can grow and divide?
Within cells, enzymes catalyse millions of chemical reactions each second. But which processes limit the speed at which cells can grow and divide?
Biologists often skirt around the ‘speed problem’ by freezing molecules in place. Given the frenzied pace of the molecular world, it is not possible to see a protein’s structure simply by training a microscope at its surface. Instead, scientists extract the proteins from cells, freeze them and then bombard them with x-rays or electrons. In this way, they can measure biological objects indirectly.
It is all the more remarkable when scientists devise an experiment that allows direct observation of a rapid biological process. Brilliant scientists have quantified many of the smallest and fastest biological processes with incredible precision, without freezing anything. Two of these experiments, in particular, left an indelible impression upon me.
In a spin
The first has to do with ATP synthase, a barrel-shaped protein, made from three subunits, that makes the energy storage molecules of the cell. ATP synthase does this by combining ADP with a free-floating phosphate to form ATP. The barrel-shaped protein sits embedded in a membrane and performs this chemical reaction by harnessing the flow of protons down a concentration gradient. It spins around and around, each rotation yielding three molecules of ATP. Making ATP is, for these reasons, both a chemical and a mechanical process.
Unfortunately, it’s not possible to watch ATP synthase spin around with one’s eyes or even a powerful microscope. The protein is too small and moves too fast. Neither can the protein be frozen in place, because then it would stop spinning entirely. More than two decades ago, though, some researchers in Tsukuba, Japan, figured out a way to get around this problem¹ and watch ATP synthase as it spins. Their experiment stands out as among the most beautiful in the history of biology.
The scientists took an ATP synthase motor and fixed it on a flat surface. They then fused one of its three subunits to a tiny gold bead. The bead was tethered to the protein via another protein, called actin, which acted as a molecular rope.
As the ATP synthase spun around, it whipped the bead in a circle. The bead measured just 40 nanometres in diameter – 50 times smaller than the length of a single Escherichia coli bacterium – but was large enough to be detected by a microscope. A camera captured an image of the rotating bead 8,000 times per second.
The scientists observed that ATP synthase spins around exactly 134 times each second, or 8,040 times each minute. That speed is significantly faster than the propeller on most piston airplanes and about half the rpm of a Boeing 737 jet engine.
Now, I can practically hear your objections through the page. “Wouldn’t the bead cause the ATP synthase motor to slow down? After all, it must add some drag!”
Yes, a bead that is too large would slow down the turbine. But the Japanese team did another experiment to ensure that wasn’t the case. They ‘tethered’ many different beads, of varying sizes, to ATP synthase. Then they repeated their experiment, carefully taking images as the differently sized beads were spun around. What they found was that, with a sufficiently small bead (such as the 40-nanometre one), it did not impede the motor’s movements whatsoever.
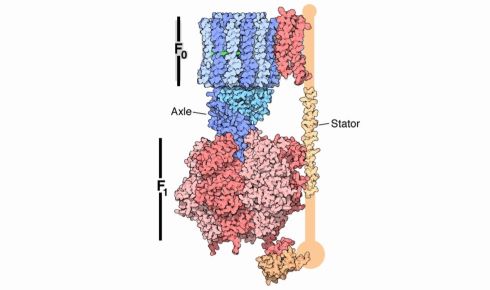 ATP synthase, which powers cellular processes, spins at 134 times per second, generating three molecules of ATP per rotation. Image courtesy of PDB-101, by the Protein Data Bank.
ATP synthase, which powers cellular processes, spins at 134 times per second, generating three molecules of ATP per rotation. Image courtesy of PDB-101, by the Protein Data Bank.
Ribosome revelation
This brings me to the second experiment. ATP is like a battery from which the cell draws energy to stay alive. Protein synthesis consumes about half of all ATP in a quickly growing bacterial cell². At any given moment thousands of messenger RNAs are being translated into proteins by ribosomes. As it moves along the mRNA strand, reads each codon and adds the corresponding amino acid to a growing protein chain, the ribosome burns ATP at each step, and all of this happens several times a second. But, again, how do we know?
In 2008 researchers at UC Berkeley devised an ingenious solution to watch individual ribosomes move along an mRNA³.
Here’s how they did it. First, the researchers chemically synthesised a strand of mRNA and attached each of its ends to a little plastic bead. Each bead was then suspended in an optical tweezer, meaning that a laser beam was directed at them and the force resulting from the scattering of photons was enough to suspend each in place.
The mRNA molecule, suspended between these beads, had been specially crafted to have a loop, or hairpin, in the middle of its sequence. The beads were held in the optical tweezers with a force of exactly 20 piconewtons – just enough to hold the mRNA molecule in place, but not enough to unwind its hairpin.
Now here's the beautiful part: as the ribosome began to move along the mRNA strand, reading its individual codons and then popping amino acids into a protein, it exerted just enough force to unwind the hairpin. And as the hairpin came undone, the mRNA stretched out in length, the two beads became more distant from one another and the increasing distance between the optical tweezers could be precisely measured.
It’s an astonishing experiment. By repeating it several times, the researchers found that ribosomes don’t glide along mRNA at a steady speed, but rather jump from one codon to the next in time steps of about 0.1 seconds. Some ribosomes even take brief pauses between each jump. We now know, through this experiment and others, that ribosomes build proteins at an average rate of 20 amino acids per second. (Recently developed methods can image single messenger RNAs, and track their movements in live cells, using microscopy⁴.)
Part of the beauty of science is that it builds upon itself, one experiment at a time. As scientists devise more experiments and take more measurements, they usually make them available in papers, preprints, websites and blogs. Then other scientists sift through these papers and compile all the numbers into an online database such as BioNumbers⁵. And now, at long last, we – humanity – have built a collection of useful numbers, a starting point, for understanding biological speed.
Sanity checks
Physicists often use order-of-magnitude calculations to do ‘sanity checks’ on their thinking.
At 5:29 on 16 July 1945, all the scientists from Los Alamos came out to watch the explosion of the first atomic bomb in the New Mexico desert. Enrico Fermi was standing about 10 miles away from the bomb and decided to estimate how much energy was released in the blast by dropping pieces of paper and seeing how much they were displaced by the wind⁶. Fermi’s estimate was accurate to within an order of magnitude. In another famous feat of estimation, Greek astronomer Eratosthenes calculated the circumference of the Earth by measuring little more than the angles of shadows. His estimate of 25,000 miles was shockingly close to the latest figure of about 24,900 miles.
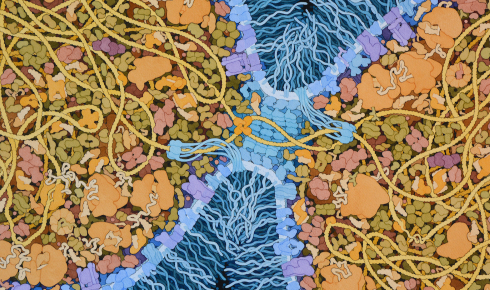 A speculative illustration of cells of the Last Universal Common Ancestor dividing, by David S Goodsell.
A speculative illustration of cells of the Last Universal Common Ancestor dividing, by David S Goodsell.
My point here is that biologists, too, can wield seemingly simple numbers to arrive at extraordinarily useful estimates. There is a surprising amount of low-hanging fruit to be gathered just from knowing the numbers involved in cellular processes, and using those numbers to perform calculations and sanity checks.
Rather than vague descriptions of a molecular phenomenon, such as DNA goes to RNA goes to protein, researchers could strive for more precision, calculating how long it would take for, say, a gene of 3,000 nucleotides to be translated into a fully folded protein. They could similarly estimate the theoretical fastest speed at which one cell could divide into two, using available numbers about the totality of atoms in a cell, the time required to copy a genome and so forth.
A recent study used order-of-magnitude estimates to correctly predict that ribosomes are the fundamental bottleneck for one cell becoming two⁷. The reason that E. coli doubles every 20 or 30 minutes, instead of every 10 minutes, is because it can’t make ribosomes fast enough. If scientists wanted to engineer cells to divide faster, then, they should start with the ribosomes.
The goal of such rough calculations is not to get the right answer, but to guide our thinking. They are a starting point for experiments, allowing one to evaluate what is possible with biology versus what ought to remain consigned to the realms of science fiction.
In my own work, I sometimes use numbers and calculations to more precisely hold an image of the cell in my mind. I can close my eyes and imagine (or at least try to imagine) what it might feel like to zoom into a bacterium, or to watch as proteins and polymerases latch onto its genome. They glide along in fits and starts, making RNA molecules at a rate of 45 nucleotides per second. Hundreds of proteins do this at the same time. The newly made RNAs jostle and push their way through a densely crowded cell. Ribosomes latch onto these RNAs, one after another, in a conga line, and begin to make protein after protein after protein, in parallel. A new protein emerges, fully formed, every 30 seconds. Proteins shuttle in and out of the cell membrane, as ATP synthases whip around and make energy storage molecules 130 times each second. It is chaotic but flowing; expertly choreographed in an often-unintelligible way.
These mental, cellular scenes are based on extant data and emerge from calculations scribbled on the backs of envelopes and scraps of paper. They help to make my imagination more vivid and precise. For when our eyes and familiar tools are insufficient to capture the true complexity of biology, we must resort to physics and mathematics. It is through numeracy that we are able to better understand the speed, and nature, of biology.
This article is an abridged version of an essay that first appeared in Asimov Press, a publisher focusing on the ideas and technologies that promote flourishing.
1) Yasuda, R. et al. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).
2) Li, G. W. et al. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157(3), 624–635 (2014).
3) Wen, J. D. et al. Following translation by single ribosomes one codon at a time. Nature 452, 598–603 (2008).
4) Katz, Z. B. et al. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife 5, 10415 (2016).
5) BioNumbers, Harvard.
6) Shapin, S. Think like a neutron. London Review of Books 40(10) (2018).
7) Belliveau, N. M. et al. Fundamental limits on the rate of bacterial growth and their influence on proteomic composition. Cell Syst. 12(9), 924–944 (2021).
Niko McCarty is a founding editor at Asimov Press


